Note
Click here to download the full example code
Plot the results of a segmentation with SPM-based pipeline T1xT2¶
# Authors: David Meunier, Bastien Cagna
# License: BSD (3-clause)
# sphinx_gallery_thumbnail_number = 2
import os
import os.path as op
import nipype.pipeline.engine as pe
from nipype.interfaces.utility import IdentityInterface
import nipype.interfaces.io as nio
from macapype.utils.utils_tests import load_test_data
Testing plot in local¶
data_path = load_test_data("data_test_macaque_processed")
# displaying results
wf_path = os.path.join(data_path, "example_segment_macaque_spm_based")
graph = os.path.join(wf_path, "graph.png")
import matplotlib.pyplot as plt # noqa
img = plt.imread(graph)
plt.figure(figsize=(36, 72))
plt.imshow(img)
plt.axis('off')
plt.show()
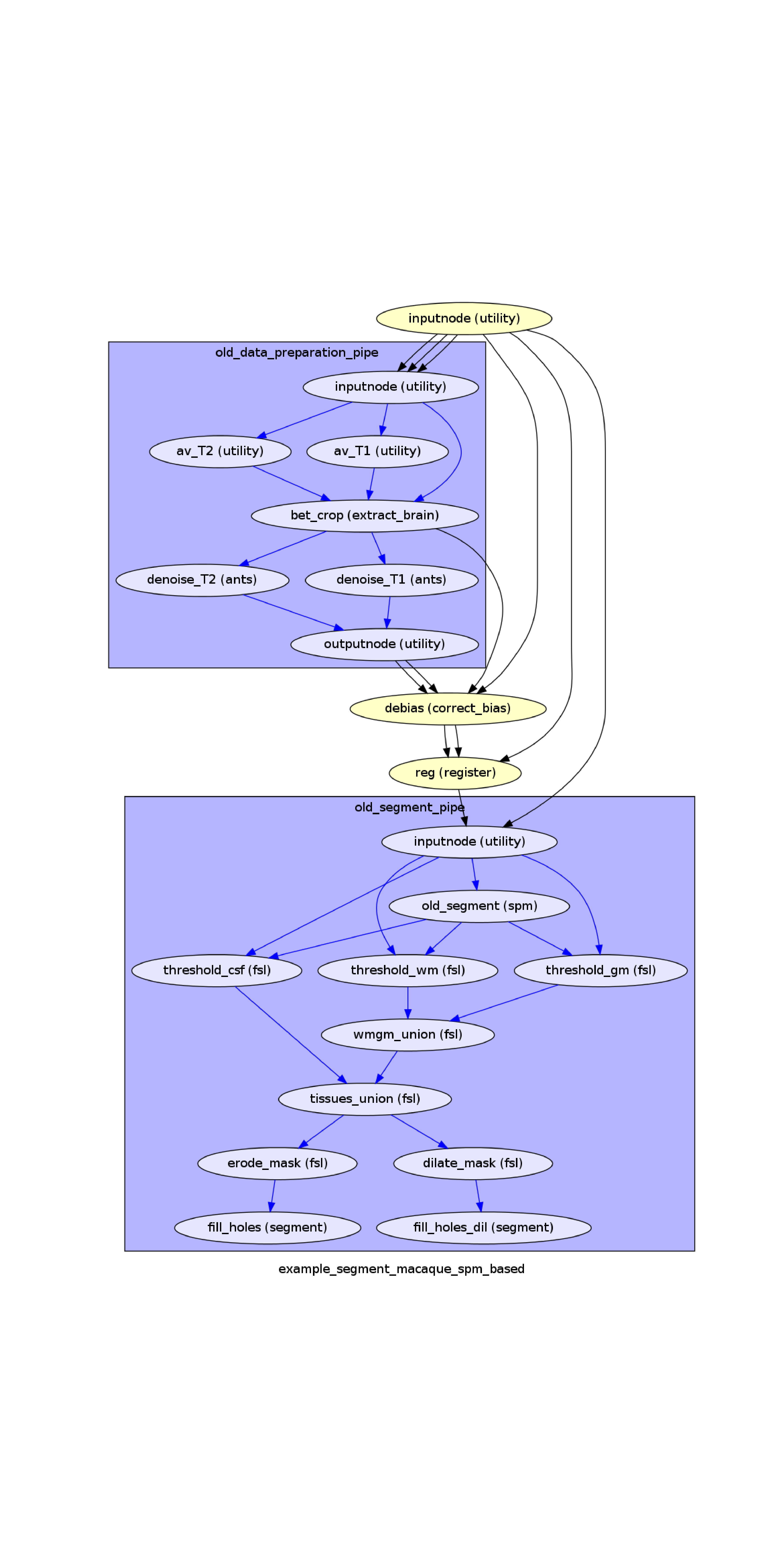
Out:
data_dirpath /home/meunier.d/data_macapype already exists
/home/meunier.d/data_macapype/data_test_macaque_processed Already exists, skipping download
Data preparation¶
results of cropping¶
cropped_T1_file = op.join(wf_path, "old_data_preparation_pipe", "bet_crop", "sub-Apache_ses-01_T1w_cropped.nii.gz")
assert op.exists(cropped_T1_file), "Error with {}".format(cropped_T1_file)
# displaying results
cropped_T1 = os.path.join(wf_path, "cropped_T1.png")
cmd = "fsleyes render --outfile {} --size 1800 600 {}".format(cropped_T1, cropped_T1_file)
os.system(cmd)
Out:
0
results of denoising¶
denoised_T1_file = os.path.join(wf_path, "old_data_preparation_pipe", "denoise_T1",
"sub-Apache_ses-01_T1w_cropped_noise_corrected.nii.gz")
assert op.exists(denoised_T1_file), "Error with {}".format(denoised_T1_file)
denoised_T1 = os.path.join(wf_path,"denoised_T1.png")
cmd = "fsleyes render --outfile {} --size 1800 600 {}".format(denoised_T1, denoised_T1_file)
os.system(cmd)
Out:
0
brain extraction results¶
bet_mask_file = op.join(wf_path, "old_data_preparation_pipe", "bet_crop", "sub-Apache_ses-01_T1w_BET_mask_cropped.nii.gz")
assert os.path.exists(bet_mask_file)
bet_mask = os.path.join(wf_path,"bet_mask.png")
cmd = "fsleyes render --outfile {} --size 1800 600 {} {} -cm blue -a 50".format(bet_mask, cropped_T1_file, bet_mask_file)
os.system(cmd)
import matplotlib.pyplot as plt # noqa
img = plt.imread(bet_mask)
plt.figure(figsize=(36, 12))
plt.imshow(img)
plt.axis('off')
plt.show()
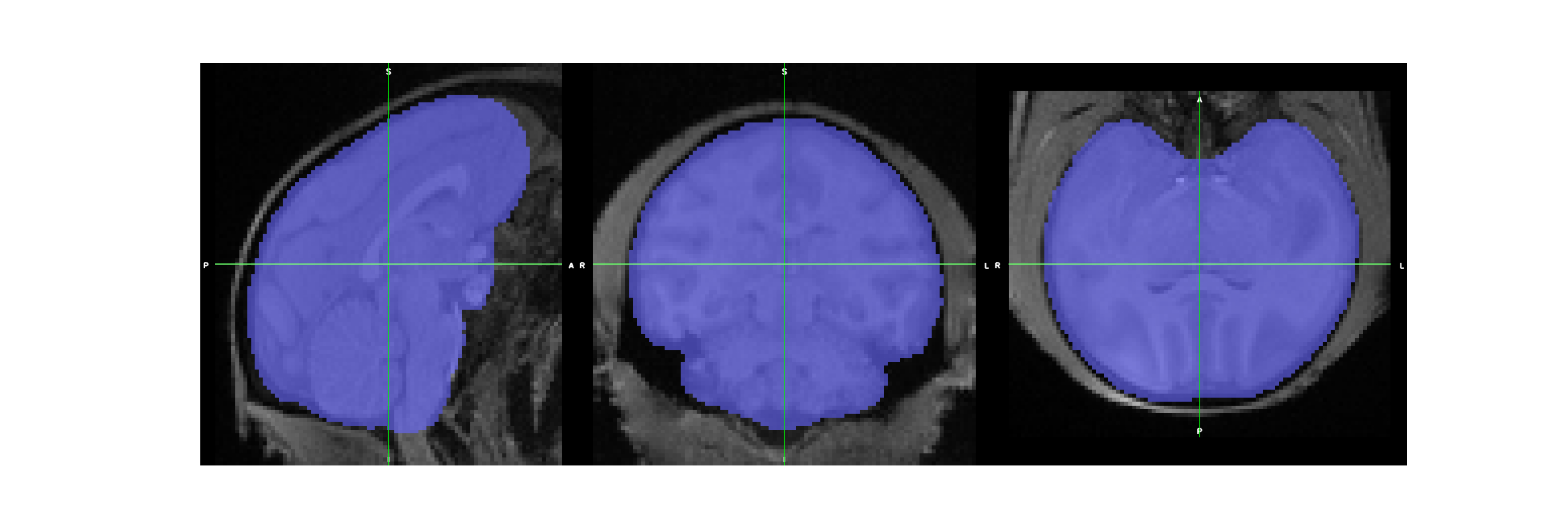
Correct bias results¶
debiased_brain_file = op.join(wf_path, "debias",
"sub-Apache_ses-01_T1w_cropped_noise_corrected_debiased_brain.nii.gz")
debiased_T1 = os.path.join(wf_path,"debiased_T1_brain.png")
cmd = "fsleyes render --outfile {} --size 1800 600 {} ".format(debiased_T1, debiased_brain_file)
os.system(cmd)
import matplotlib.pyplot as plt # noqa
fig, axs = plt.subplots(3, 1, figsize=(36, 36))
axs[0].imshow(plt.imread(cropped_T1))
axs[0].axis('off')
axs[1].imshow(plt.imread(denoised_T1))
axs[1].axis('off')
axs[2].imshow(plt.imread(debiased_T1))
axs[2].axis('off')
plt.show()
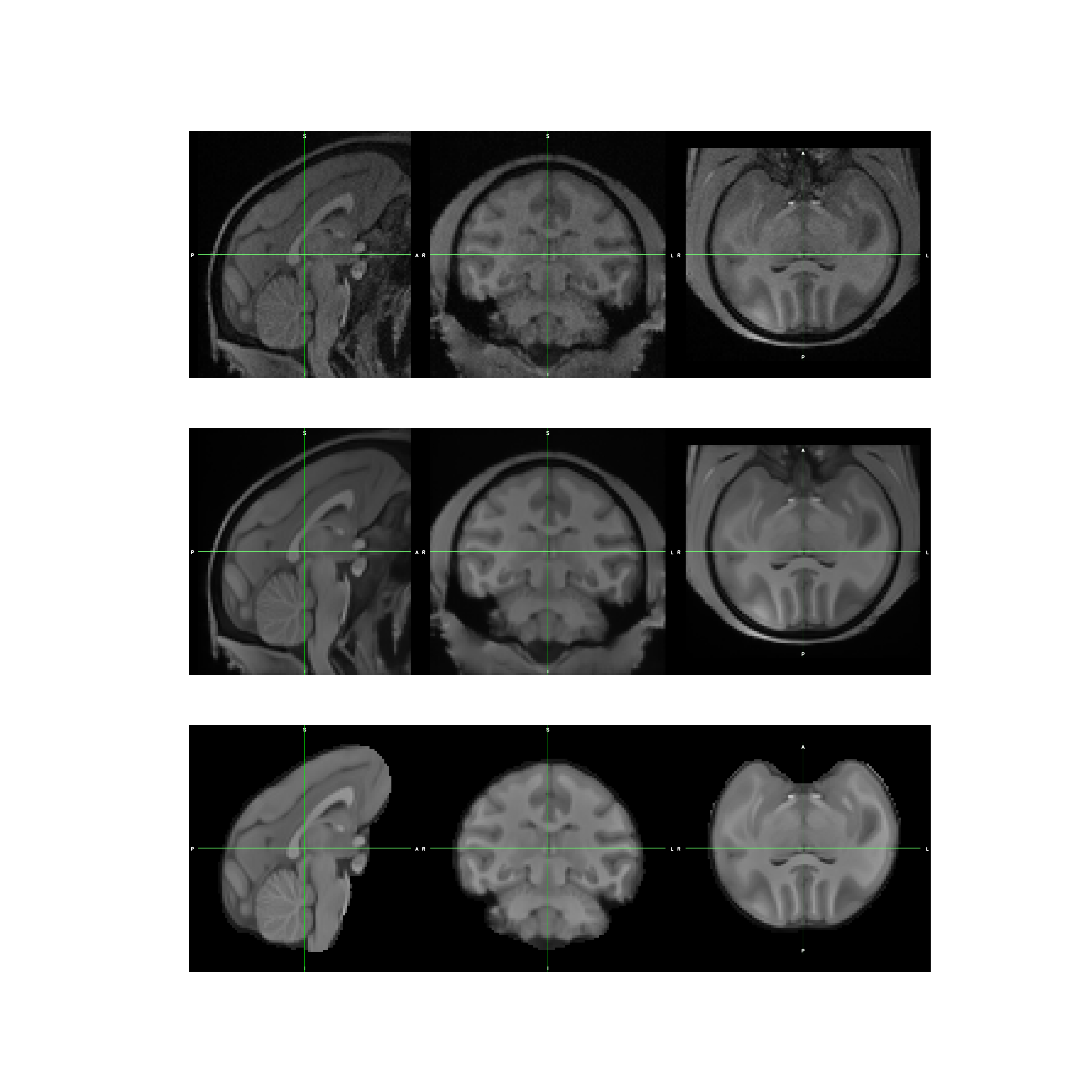
registration results¶
reg_T1_file = op.join(wf_path,"reg", "sub-Apache_ses-01_T1w_cropped_noise_corrected_debiased_brain_FLIRT-to_inia19-t1-brain.nii")
assert os.path.exists(reg_T1_file)
# showing mask
segment_path = os.path.join(wf_path, "old_segment_pipe")
filled_mask_file = os.path.join(segment_path, "fill_holes", "c1sub-Apache_ses-01_T1w_cropped_noise_corrected_debiased_brain_FLIRT-to_inia19-t1-brain_thresh_maths_maths_ero_filled.nii.gz")
assert os.path.exists(filled_mask_file)
#filled_mask_file = os.path.join(segment_path, "fill_holes_dil", "c1sub-Apache_ses-01_T1w_cropped_debiased_brain_FLIRT-to_inia19-t1-brain_thresh_maths_maths_dil_filled.nii.gz")
outfile_seg_mask = os.path.join(wf_path,"outfile_seg_mask.png")
cmd = "fsleyes render --outfile {} --size 1800 600 {} {} -a 50".format(outfile_seg_mask, reg_T1_file, filled_mask_file)
os.system(cmd)
import matplotlib.pyplot as plt # noqa
img = plt.imread(outfile_seg_mask)
plt.figure(figsize=(36, 12))
plt.imshow(img)
plt.axis('off')
plt.show()
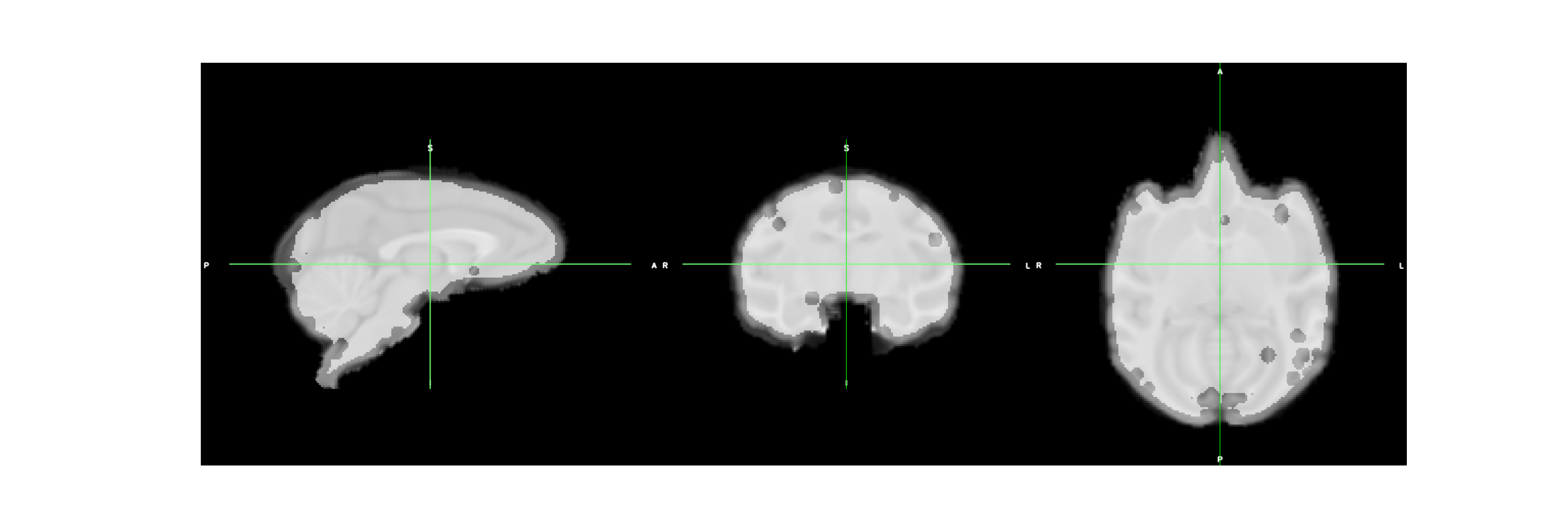
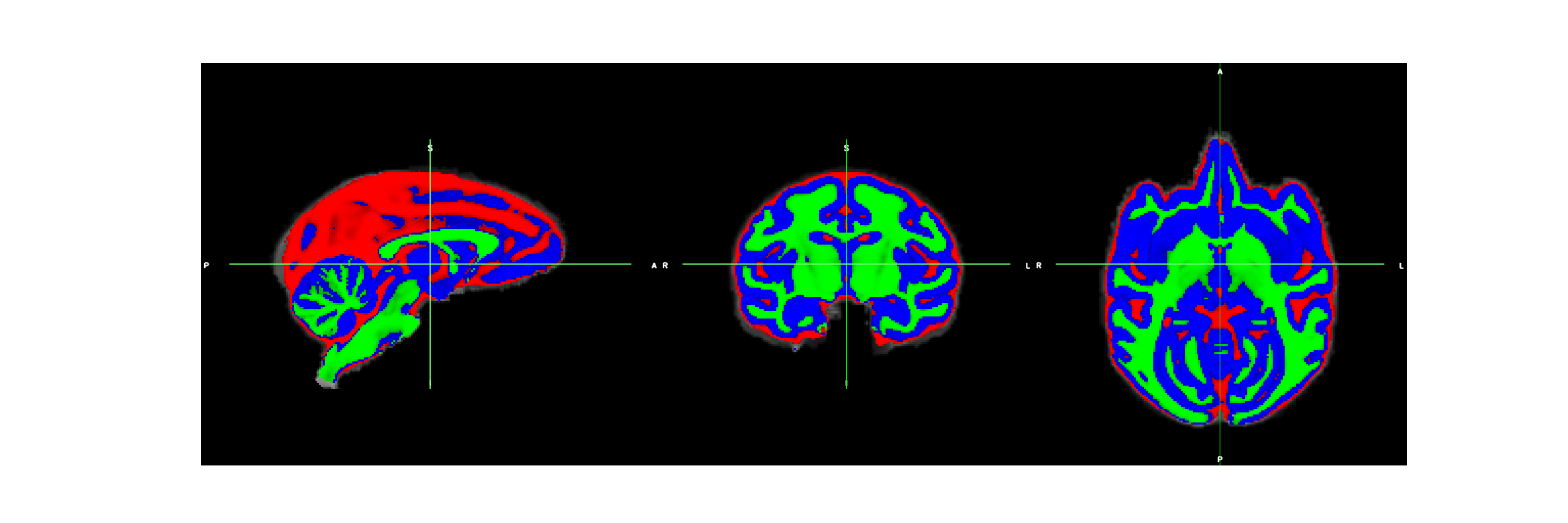
segmentation results¶
gm_file = os.path.join(segment_path, "threshold_gm", "c1sub-Apache_ses-01_T1w_cropped_noise_corrected_debiased_brain_FLIRT-to_inia19-t1-brain_thresh.nii.gz")
wm_file = os.path.join(segment_path, "threshold_wm", "c2sub-Apache_ses-01_T1w_cropped_noise_corrected_debiased_brain_FLIRT-to_inia19-t1-brain_thresh.nii.gz")
csf_file = os.path.join(segment_path, "threshold_csf", "c3sub-Apache_ses-01_T1w_cropped_noise_corrected_debiased_brain_FLIRT-to_inia19-t1-brain_thresh.nii.gz")
assert os.path.exists(gm_file)
assert os.path.exists(wm_file)
assert os.path.exists(csf_file)
outfile_seg_col = os.path.join(wf_path,"outfile_seg_col.png")
cmd = "fsleyes render --outfile {} --size 1800 600 {} {} {} {}".format(outfile_seg_col, reg_T1_file, gm_file, wm_file, csf_file)
cmd = "fsleyes render --outfile {} --size 1800 600 {} {} -cm red {} -cm blue {} -cm green".format(outfile_seg_col, reg_T1_file, gm_file, wm_file, csf_file)
os.system(cmd)
import matplotlib.pyplot as plt # noqa
img = plt.imread(outfile_seg_col)
plt.figure(figsize=(36, 12))
plt.imshow(img)
plt.axis('off')
plt.show()
Total running time of the script: ( 0 minutes 57.736 seconds)
